Due: Fri Feb 26th, 11:59PM EST
100 points total
Please read: Assignment 3 Advice, Tips, and Clarifications
Overview
In this assignment you will design a simple C++ framework that makes it surprisingly simple to create high-performance, parallel implementations of a number of graph processing algorithms. Since your implementation will run in parallel on the 61 cores (actually only 59 are available for our applications) of a Intel Xeon Phi processor, and since it's a system for processing graphs, we've given the framework the name... ParaGraph! A good implementation of this assignment will be able to run algorithms like breadth-first search on graphs containing hundreds of millions of edges in just over a second.
Environment Setup
In this assignment you will be writing code in C for the 61 core Xeon Phi 5110p processors in the latedays.andrew.cmu.edu cluster. Details on these chips is available here. Each core has 4-way multi-threading.
On latedays, all jobs are run through a job queue. More details on how to run jobs on latedays is described here.
To get started:
Add the following to your .bashrc on
latedays.andrew.cmu.edu:export PATH="/opt/intel/bin:${PATH}";source /opt/intel/bin/compilervars.sh intel64As you did for previous assignments, download the Assignment 3 starter code from the course Github using:
git clone https://github.com/cmu15418/assignment3
This assignment will be written in C++. For any C++ questions (like what does the virtual keyword mean), the C++ Super-FAQ is a great resource that explains things in a way that's detailed yet easy to understand (unlike a lot of C++ resources), and was co-written by Bjarne Stroustrup, the creator of C++!
Background
Representing Directed Graphs
The ParaGraph framework operates on directed graphs, whose implementation you can find in graph_internal.h. A graph is represented by an array of edges (both outgoing_edges and incoming_edges), where each edge is represented by an integer describing the id of the destination vertex. Edges are stored in the graph sorted by their source vertex, so the source vertex is implicit in the representation. This makes for a compact representation of the graph, and also allows it to be stored contiguously in memory. For example, to iterate over the outgoing edges for all nodes in the graph, you'd use the following code which makes use of convenient helper functions defined in graph.h (and implemented in graph_internal.h):
for (int i=0; i<num_nodes(g); i++) {
// Vertex is typedef'ed to an int. Vertex* points into g.outgoing_edges[]
const Vertex* start = outgoing_begin(g, i);
const Vertex* end = outgoing_end(g, i);
for (const Vertex* v=start; v!=end; v++)
printf("Edge %u %u\n", i, *v);
}
ParaGraph's Two Graph Operators: VertexMap and EdgeMap
The ParaGraph framework provides applications with two operations for processing graphs: vertexMap, and edgeMap. These operations, as indicated by their name, map an application-provided function onto edges and vertices of the graph.
vertexMap is defined (abstractly) as follows:
vertexMap(U: vertexSubset,
F: vertex -> bool) : vertexSubset
1. apply F to all vertices in U
2. return a new vertex subset containing all vertices u in U
for which F(u) == true
In other words, vertexMap accepts as input a set of vertices from the graph, and a function that, given a single vertex, returns the value true or false. It applies the function F to all vertices in the set U, and returns a new set containing only the vertices u for which F(u) evaluates to true.
edgeMap is a bit more complex, and is defined as follows:
edgeMap(G: graph,
U: vertexSubset,
C: vertex -> bool,
F: (vertex, vertex) -> bool) : vertexSubset
1. Apply F to all edges (u,v), where u is in U and v satisfies the condition C(v)
2. return a new vertex subset containing all vertices v
for which F(u,v) == true
In other words edgeMap accepts as input a set of vertices from the graph and two functions:
- A condition function
Cthat accepts a single input vertex and returnstrueorfalse. This function is used to filter the set of edges onto whichFis applied. - A function
Fthat takes as input an edge (u,v) from the graph and returnstrueorfalse.Fis mapped onto all edges (u,v) whose source vertexuis inUand whose destination vertexvpasses the condition functionC.
edgeMap returns a new vertexSubset that contains all destination vertices v for which F(u,v) == true. Note that since F may evaluate to true for multiple graph edges that point to v, care must be taken to not include v into the output vertex set twice (it is a set after all). More detailed pseudocode for edgeMap is given below:
edgeMap(G: graph,
U: vertexSubset,
C: vertex -> bool,
F: (vertex, vertex) -> bool) : vertexSubset
outputSubset = {}
foreach u in U: (in parallel)
for each outgoing edge (u,v) from u: (in parallel)
if (C(v) && F(u,v))
outputSubset.append(v)
remove_duplicates(outputSubset)
return outputSubset
edgeMap is an interesting operator because it doesn't operate on a collection, it interprets the structure of a graph. However, with just vertexMap and edgeMap, you can implement a surprisingly wide set of graph algorithms, for example, consider the parallel implementation of breadth-first search, implemented in a much more C-like syntax below:
int* parents = ... initialied to -1
bool cond(int v) {
return parents[v] == -1;
}
bool update(int u, int v) {
return compare_and_swap(& parents[v], -1, u);
}
void bfs(graph g, int root, int* parents) {
parents[root] = root; // parent of root vertex is itself
vertexSubset frontier = {root};
while (size(frontier) > 0) {
frontier = edgeMap(graph, frontier, cond, update);
}
}
There are two important details to note about this code:
- The function
Fis allowed to have side-effects (here it updates the arrayparents), and so the implementation ofFmust take care that it is correct even in the face of parallel execution. - The BFS example above uses efficient
compare_and_swapoperation that performs an atomic conditional read-modify-write operation on the address given in its first parameter. You can read about GCC's implementation of compare and swap, which is exposed to C code as the function__sync_bool_compare_and_swap. (We will talk about compare and swap in detail in the second half of the course, but in this problem it's up to you to figure out how to use it.) Consider an implementation ofupdatewithout the call to 'compare_and_swap`. How will the frontier be different if this synchronization was not added.
Part 1: What You Need to Do
In part one of this assignment you need to a provide correct parallel implementation of the ParaGraph framework using OpenMP for parallelism. Specifically, this means you need to:
- Provide implementations of
edgeMapandvertexMapinparaGraph.h - Provide an implementation of
VertexSetinvertex_set.handvertex_set.cpp
When you have completed this part of the assignment, your implementation will be able to successfully run the applications in the apps directory. This includes BFS as well as:
kbfs: A program that estimates the radius (eccentricity) of the input graph using a modified BFS algorithm that simultaneously performskBFS's starting akrandom nodes in the graph. The radius of a graph is the longest path in the graph between two vertices.pagerank: The basic Google page rank algorithm for computing the relative importance of web pages (graph vertices) given the topology of links between these pages (graph edges).
It's quite interesting that these very different algorithms can be implemented using just edgeMap and vertexMap operations!
Note that your implementation should be correct at this point, but it may not be particularly fast compared to the reference.
How to Compile and Run
While your implementation's performance will be assessed on the Xeon Phi Processor, to help you debug your program, you also have the option to compile to CPU instead of the Phi. To compile and execute for CPU, run:
make cpu
./paraGraph <app> <path to graph>
For example:
./paraGraph bfs /home/15-418/soc-slashdot_900k.graph
HOWEVER, PLEASE, PLEASE DO NOT RUN LARGE TEST RUNS ON THE HEAD NODE OF LATEDAYS.ANDREW.CMU.EDU. Remember, everyone is logged into this node and probably using an interactive shell editing their code. If you run your code on big graphs on the head node, you'll slow it down for everyone. If you want to test your code on the CPU, please use the gates machines.
If you want to run on large graphs, or on the Xeon Phi, you should use the latedays job queue. You can submit your job to the queue using the following command:
make clean
make APP="app" GRAPH="path to graph" jobs
qsub jobs/${USER}_${APP}_${GRAPH}.job
You can monitor the progress of your job by running:
qstat -u ${USER}
Note that you can access AFS from latedays head node by using the symlink in your home directory (~/AFS). However, when you submit your job to the job queue, the worker nodes don't have access to AFS. Therefore, you should compile your assignment on the latedays filesystem and submit that copy to the job queue.
The app argument supports the following:
bfs
kbfs
pagerank
decomp
grade # the grading harness runs bfs, kbfs, and pagerank
You can use the -t flag to control the number of threads you want use.
Here's a sample output of runnings bfs on soc-slashdot_900k.graph:
------------------------------------------------------------------------------
Running on device 0
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Max system threads = 24
Running with 24 threads
------------------------------------------------------------------------------
Loading graph...
Graph stats:
Nodes: 82168
Edges: 948464
Timing results for BFS:
===========================================================================
Threads Ref. Time Ref. Speedup Your Time Your Speedup
---------------------------------------------------------------------------
1 0.0035 1.0000 1.0000 1.0000
2 0.0024 1.4397 1.0000 1.0000
4 0.0017 2.0840 1.0000 1.0000
8 0.0016 2.2300 1.0000 1.0000
16 0.0017 2.0216 1.0000 1.0000
24 0.0225 0.1540 1.0000 1.0000
---------------------------------------------------------------------------
Time: 64307.91% of reference solution. Grade: 0.00
---------------------------------------------------------------------------
Total Grade: 0.00/4.50
Notice that the reference solution performs horribly with 24 threads! This may be due to the fact that the graph is small, so there is a limited amount of parallelism that we can exploit.
Where to find testing graphs
On latedays, graphs can be found at /home/15-418/asst3_graphs. On Andrew Linux, graphs can be found at /afs/cs/academic/class/15418-s16/public/asst3_graphs/. Note that some of these graphs are quite big (> 2GB) so it's likely you won't want to localize them into your home directory.
How to Test
You will be testing your implementation of ParaGraph on BFS, kBFS, and PageRank. For more detail on how these algorithms work, seee the documentation in the starter code. For kBFS you can use either the compareArraysAndDisplay or compareArraysAndRadiiEst test functions in main.cpp.
Note that compareArraysAndDisplay will print out the radii estimates for each node in the graph. You should only use this for the small grid graphs. Use compareArraysAndRadiiEst for testing kBFS correctness on larger graphs.
A Few Tips and Hints:
- In addition to implementing the logic of
edgeMapandvertexMap, an important part of this part of the assignment is choosing how to representvertexSubsets.
How to Test Performance
Once you're ready to test the performance of your code, run
make grade
qsub jobs/${USER}_grade_performance.job
Learning OpenMP
OpenMP is an API and set of C-language extensions that provides compiler support for parallelism. It is well documented online, but here is a brief example of parallelizing a for loop, with mutual exclusion
You can also use OpenMP to tell the compiler to parallelize iterations of for loops, and to manage mutual exclusion.
/* The iterations this for loop may be parallelized */
#pragma omp parallel for
for (int i = 0; i < 100; i++) {
/* different iterations of this part of the loop body may be
run in parallel on different cores */
#pragma omp critical
{
/* This block will be executed by at most one thread at a time. */
printf("Thread %d got iteration %lu\n", omp_get_thread_num(), i);
}
}
Please see OpenMP documentation for the syntax for how to tell OpenMP to use different forms of static or dynamic scheduling (e.g., omp parallel for schedule(dynamic)).
Here is an example for an atomic counter update.
int my_counter = 0;
#pragma omp parallel for
for (int i = 0; i < 100; i++) {
if ( ... some condition ...) {
#pragma omp atomic
my_counter++;
}
}
As a participant in a 400-level course, we expect you to be able to read OpenMP documentation on your own (Google will be very helpful), but here are some useful links to get started:
- The OpenMP 3.0 specification: http://www.openmp.org/mp-documents/spec30.pdf.
- An OpenMP cheat sheet http://openmp.org/mp-documents/OpenMP3.1-CCard.pdf.
- OpenMP has support for reductions on shared variables, and for declaring thread-local copies of variables.
Part 2: Another Way to Implement EdgeMap
If you take a closer look at the step-by-step results of calls to edgeMap in BFS, you'll notice that the frontier grows quite large in some steps (sometimes growing to contain a large fraction of the entire graph). Think about why this behavior might cause a performance problem in the BFS implementation from Part 1. This suggests an alternative implementation of a breadth-first search step that may be more efficient in these situations. Instead of iterating over all vertices in the frontier and marking all vertices adjacent to the frontier, it is possible to implement BFS by having each vertex check whether it should be added to the frontier! Basic pseudocode for the algorithm is as follows:
for each vertex v in graph:
if v has not been visited AND
v shares an incoming edge with a vertex u on the frontier:
add vertex v to frontier;
This algorithm is sometimes referred to as a "bottom up" implementation of BFS, since each vertex looks "up the BFS tree" to find its ancestor. (As opposed to being found by its ancestor in a "top down" fashion, as done in Part 1.) Without changing the implementation of BFS at all, you can change your implementation of edgeMap to implement this bottom up BFS. Start by implementing a simple sequential version of this modified edgeMap, then parallelize your implementation and compare the performance of "down top" and "bottom up" implementations. More specifically:
Notice that in some steps of the BFS, the "bottom up" BFS is signficantly faster than the top-down version. On other steps, the top-down version is signficantly faster. This suggests a major performance improvement in your implementation of edgeMap, you could dynamically choose between your "top down" and "bottom up" formulations based on the size of the vertexSubset or other properties of the graph! If you want a solution competitive with the reference, your implementation will likely have to implement this dynamic optimization.
With this new implementation of edgeMap, you have all the tools to be performance competitive with the reference implementation.
A Few Tips and Hints:
Think about how you can express bottom-up BFS in terms of the
CandFfunctions that are supplied toedgeMap. More specifically:- "if v has not been visited" corresponds checking the result of the
cond()function. - "if v shares an incoming edge edge a vertex u on the frontier" corresponds to checking the result of F(u,v)
- Last, note that in BFS
vis added to the new frontier if there's ANY edge from a vertex on the frontier to v. Or equivalently, if evaluating F(u,v) causes cond(v) to become true. How can you exploit this in your implementation? (You should also take a look at how each app defines theCandFfunctions and see if there's any property that you can exploit. )
- "if v has not been visited" corresponds checking the result of the
At this point in the problem it may be useful to consider how to represent
vertexSubsetwhen you are using the bottom up and top down versions ofedgeMap. For example, avertexSubsetthat contains a large fraction of the nodes of a graph might be best represented differently than the set of vertiex ids you likely used in Part 1.- This is a good time to take a harder took at the
compare_and_swapused in the BFS application.compare_and_swapwill be treated by the cache coherence protocol as a write (even if the compare fails). Is there a way to avoid thecompare_and_swapin some situations? - Note that the bottom-up
edgeMapapproach is not necessarily faster for all apps! - If you have memory errors (segfault, double free, etc.), use
valgrind, it'll tell you exactly what line they occur on! Quick start instructions are at http://valgrind.org/docs/manual/quick-start.html.
Part 3: Implementing Parallel Graph Decomposition
One sign of a well-designed system is that only a few operations are needed to efficiently express a wide variey of use cases. So far in this assignment, you've seen how the edgeMap and vertexMap primitives are sufficiently powerful to express BFS, kBFS, and a variant of pageRank. In the final part of the assignment we'll ask you to implement one additional application: graph decomposition.
Several times in class we've described situations where it was important to decompose a dataset into pieces that can be processed in parallel, while also choosing a decomposition strategy that maintains high data locality (reduces communication). In this part of the assignment you'll implement a new algorithm that computes a decomposition of a graph with some nice locality properties.
The algorithm, which was recently developed by Professor Miller and students here at CMU, is called Graph Decomposition Using Random Shifts. All the information needed to implement the algorithm is below, but for those interested, you can read the full paper on the algorithm here.
The goal of this algorithm is to decompose an undirected unweighted graph into subgraphs that have small diameter (the number of steps between two nodes in the same subgraph) and where the fraction of edges connecting nodes in different subgraphs is small. In other words, you want to partition the graph into many small subgraphs with high locality. Although in this assignment we won't use this decomposition in the context of a larger graph-processing application, you can imagine how graph partitionings could be a useful sub-routine in more complex graph algorithms, or for parallelizing graph-based algorithms across a cluster.
More specifically, given an undirected, unweighted graph G = (V,E) with m edges and n nodes, a (beta, d) decomposition is a partition of V into subsets S_i ... S_k such that:
- The strong diameter of each
S_iis at mostd. The strong diameter of a subgraph is the largest distance between its vertices. - The number of edges with endpoints belonging to different pieces is at most
beta*m.
The Miller et al. paper describes how a standard choice of parameters for a decomposition is (beta,O(log(n)/beta)).
One way to cluster a graph sequentially is to use a method called "ball growing". Ball growing starts with single randomly chosen vertex in the graph, and repeatedly adds the neighbors of the current set into an "active set" using a breadth-first traversal of the graph from this starting vertex. The traversal terminates when the number of edges on the boundary of the active set is less than a beta fraction of the edges within the "ball" (the subgraph of vertices and edges traversed so far). Once this first cluster is found, the algorithm discards all vertices from the first "ball", repeats the ball growing process on a new random node in the remaining graph. Notice that in the sequential algorithm we cannot grow the next ball until the current ball has finished growing.
To perform decomposition in parallel, we will instead employ an approach that is similar to in spirit to performing multiple BFS searches in parallel, except now BFS searches will start from graph nodes at random times during the search process. The first search that touches a node in the graph will "claim" that vertex for the subgraph associated with the root vertex of the BFS. In other words, the algorithm will perform many BFS's in parallel, but gradually increase the number of BFS's simultaneously being performed in order to make sure no one subgraph gets too large.
At the beginning of the algorithm, each vertex i in the graph is a potential ball center and is assigned a random value deltamu_i generated from an exponential distribution with rate 1/beta. (See function getDus in the starter code file apps/graph_decomposition.cpp). The time (i.e. iteration) at which this ball center can begin to grow is determined by its deltamu. The larger the deltamu, the earlier it can begin to grow. Furthermore, if a potential ball center has already been claimed by some other ball by the time it is allowed to start growing, the BFS for this vertex is skipped (it is already part of some other subgraph). The first ball to reach a vertex is the owner of that vertex.
Below in pseudocode are the steps of the algorithm:
// Given a graph, a list of exponentially generated delta mus (cast to int),
// the maximum delta mu value, and the id of the vertex that posseses the maximum
// delta mu value, perform the parallel graph decomposition
void decompose(graph *g, int *decomp, int* dus, int maxDu, int maxId) {
VertexSet* frontier = newVertexSet(1);
frontier->addVertex(maxId); // vertex with maxDu grows first
iter = 0;
while (frontier->size > 0) {
foreach src vertex on frontier {
foreach dest vertex of src {
if (previously_unclaimed(dest) && min_candidate_owner(src.ball)) {
dest.ball = src.ball // mark dest as claimed by ball of src vertex
new_frontier->addVertex(i);
}
}
}
iter++;
// start growing all balls i at the next iter with
// unvisited center i and with maxDu - dus[i] < iter
foreach vertex i {
if (unclaimed(i) && iter > du_max - dus[i]) {
new_frontier->addVertex(i);
}
}
frontier = new_frontier;
}
}
previously_unclaimed(dest) is true if the destination vertex was unclaimed at the start of this iteration. min_candidate_owner(src.ball) will evaluate to true if the ball that source belongs to has the smallest id seen so far out of all the balls that have attempted to claim the destination vertex on this iteration.
Here's a concrete example: say two balls, one originating at vertex 7 and another at vertex 12 both reach an unvisited vertex 24 on the ith iteration. How do we decide which ball to give vertex 24 to? A correct implementation will compare the ids of both balls and assign vertex 24 to the ball with the smaller center id. So in this case, that would be the ball centered at vertex 7. Keep in mind that it is possible for two or more vertices to attempt to edit the cluster id of a given vertex simultaneously or at different points in time within the same iteration. You'll have to make sure your implementations makes sure that in such cases, the destination vertex gets assigned to the correct ball.
In the loop over vertices at the end of the in the code above, unclaimed(i) will evaluate to true if vertex i is currently unclaimed.
What to need to do:
Implement decompose(graph* g, int *decomp, int* dus, int maxVal, int maxId) in apps/graph_decomposition.cpp. Given a graph, a list of exponentially generated du's (cast to ints), the maximum du value, and the id of the vertex that posseses the maximum du value, decompose performs the parallel graph decomposition algorithm described above and stores the id of the ball that vertex i belongs to in decomp[i].
A Few Tips and Hints:
You will have to maintain a list of cluster ids where cluster[i] is the cluster that vertex i belongs to. The id of a cluster is just the id of the vertex at its center. If you plan on using this list to check whether or not a vertex has been already claimed by a cluster, be very careful about how updating this list within an iteration of the algorithm could affect the correctness of your implementation, specifically in cases where two or more different source vertices attempt to claim a destination vertex on the same iteration.
How to test your code:
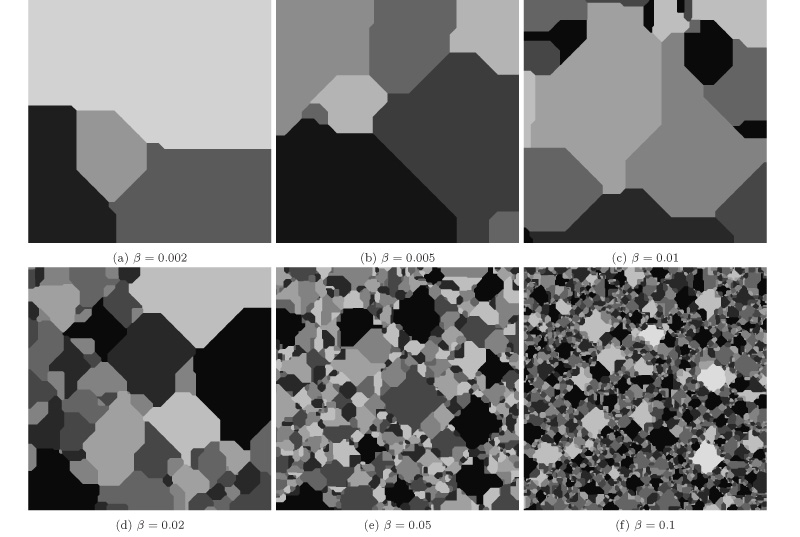
We recommend testing your code on the grid graphs. When testing, you can use the compareArraysAndDisplay test function inside main.cpp to visualize what your graph decomposition looks like. This function will print out the cluster id that each node in the grid belongs to. Use this for the smaller grids. For larger grids we recommend just using the compareArrays function inside main.cpp. Note the DecompBeta value defined at the top of main.cpp. Larger values of beta generate smaller clusters and vice versa. A reasonable value for the smaller graphs is 2 so that you can actually see clusters instead of one big cluster. Feel free to play around with different values of beta. Your clusterings should look similar to those shown in the above images as you change your beta value.
Grading
- (15 points) ParaGraph Runtime Correctness. A correct implementation of
edgeMapandvertexMapthat is able to run all applications on the graphs used in the grading script. Final correctness testing may be performed on additional graphs as well - (54 points) Performance. Your performance score will be the sum of your performance scores on 12 test configurations (3 applications and 4 graphs for each application). It is not expected that you optimize your implementation per application, so the wide test matrix largely serves to compute an average overall performance score. Performance is graded using your best wall-clock time (not speedup) after running the code in 64, 128, and 240 thread configurations. The performance testing configurations are:
- Applications:
BFS,kBFS,pagerank - Graphs:
soc-pokec_30m.graph,rmat_200m.graph,soc-livejournal1_68m.graph,com-orkut_117m.graph - Each configutation is worth 4.5 points
- A solution less than 1/4 the performance of the reference is 0 points
- A solution matching the reference's performance is 4.5 points
- Applications:
- (6 points) Graph Decomposition Correctness. A correct implementation of the graph decomposition algorithm is worth 6 points.
(5 points) Graph Decomposition Performance. A solution matching the reference's performance is 5 points.
(20 points) Writeup. Your write-up should contain a detailed description your optimization process and we will grade the writeup on the quality of this description. Make sure you tell us how you implemented (and parallelized)
vertexMapandedgeMap, as well as how you chose to representvertexSubsets. In addition to your algorithm description, we are looking for you to analyze the performance of your program. Specifically address the following questions:- Where is the synchronization in your solution? Did you do anything to limit the overhead of synchronization?
- Why do you think your code (and the TA's reference code) is unable to achieve perfect speedup? (Is it workload imbalance? communication/synchronization? data movement?)
- At high thread counts, do you observe a drop-off in performance? If so, (and you may not) why do you think this might be the case?
- If you modified the apps, please describe the modifications and the reason behind your change.
Hand-in Instructions
make handin and submit the resulting tar file on Autolab. Please note that only files you are supposed to change are submitted. Any other files or changes will be lost. Make sure all of your code is in paraGraph.h, vertex_set.h, vertex_set.cpp, or in one of the app cpp files (i.e. app/*.cpp). Name your writeup writeup.pdf.
Please note, you will definitely have to change paraGraph.h, vertex_set.h, vertex_set.cpp, and apps/graph_decomposition.cpp. While it's not strictly required, it may be also be good to optimize the other applications at apps/*.cpp.